
In this calculation frequencies are directly inferred as per the definition of the HW equation. The second method of utilising the HW equation is a lot more direct, and just about every text book I could find uses this method to solve 3+ allele problems (as, for example, blood types) Wikipedia commends its usefulness in calculating heterozygote frequencies of genetic diseases in large populations. Using these values for p and q in the HW equation yields:Īnd a population-wide phenotype distribution of Phenotype n Phenotype n | HW1 HW2p HW2qĪs suggested by above-linked data set source and Beebee/Rowe's Introduction to Molecular Ecology, alleles are simply added up to get values for p and q respectively: Phenotype n | P Qįrom this we can directly infer p/q: $p = 94/(94+34) =0.73$ and $q = 34/(94+34) = 0.27$ with the condition $p + q = 1$ remaining true. I'm using observed phenotype in the place of the original data's observed genotype. is it valid to use method #1 on 3-allele-data sets)? Or do I just use whatever method is easiest? Doesn't this lead to discrepancies in the results? This seems like a big thing, especially when comparing real-world observation data to assumed HW distributions.Įxample data set (EST/California) and results of HW calculations. Is either of these methods (technically) wrong? How do I decide which one to use for each data set (i.e. I feel like I'm not seeing the forest for the trees, to be honest. There are two different methods of working with the HW equation that I have come across, and they do not seem to be compatible used on the same data sets they yield wildly different results.
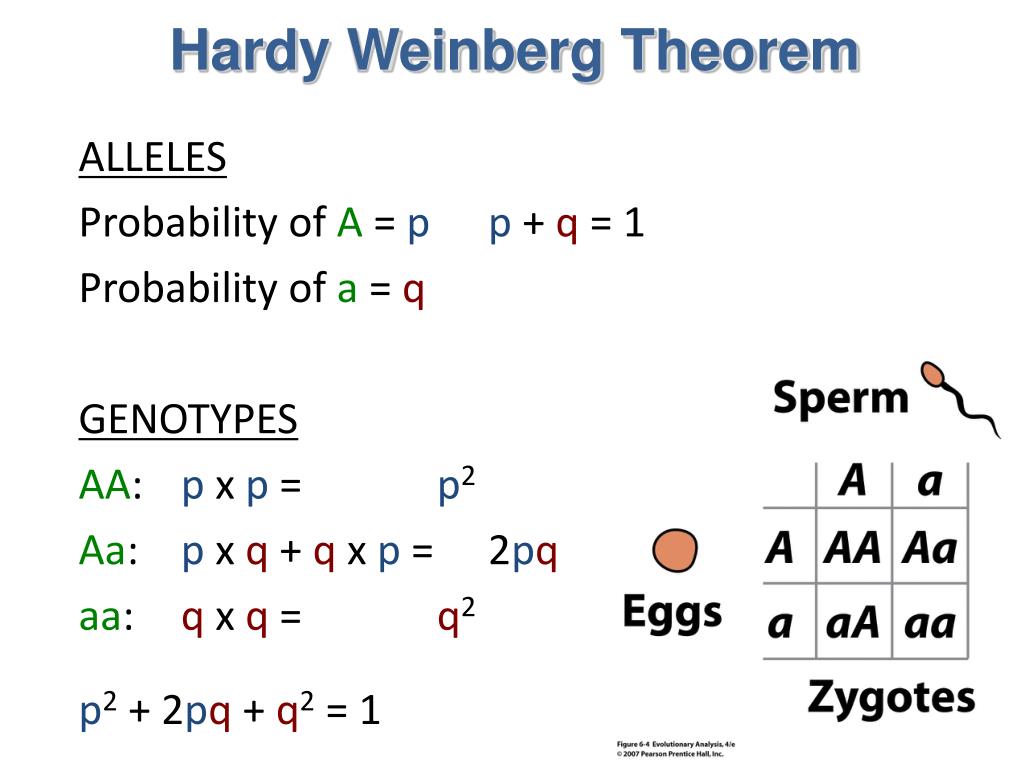
$q^2$ = Frequency of homozygous phenotype QQ $2pq$ = Frequency of heterozygous phenotype PQ $p^2$ = Frequency of homozygous phenotype PP The formula's theory assumes a binomal distribution of allele frequencies in a population, and hence allows the comparison of observed phenotype frequencies to an ideal model distribution. This breaks down to 10 homozygous genotypes and 45 heterozygous genotypes.I am more than flabbergasted by calculations of Hardy-Weinberg equilibria. If a population has 10 alleles for a specific gene, the combined, total number of homozygous and heterozygous genotypes present in the population will be: However, the equation above can be used to calculate the number of genotypes for a locus with any number alleles. The calculator does not go beyond 5 alleles and 15 possible genotypes. The general formula for finding the sum of a set of integers from 1 to n is:

With 2 alleles, the number of genotypes is 1 + 2 = 3.Given n alleles at a locus, the number genotypes possible is the sum of the integers between 1 and n: Number of genotypes for a given number of alleles To avoid having your values changed, make sure your values sum to one and enter them from top to bottoms (p then q then r. The calculator has a check that prevents the allele frequencies from summing to any value other than 1.
Hardy weinberg equation calculator update#
Update the values by changing the allele frequency in the blue box below the graph.


 0 kommentar(er)
0 kommentar(er)
